Author
Author  Correspondence author
Correspondence author
Triticeae Genomics and Genetics, 2010, Vol. 1, No. 1 doi: 10.5376/tgg.2010.01.0001
Received: 04 Apr., 2009 Accepted: 27 Apr., 2009 Published: 28 May, 2009
Li et al., 2009, RAPD Analysis on Variation of Mitochondrial DNA for Cytoplasmic-Nuclear Male Sterile Lines in Wheat, Molecular Plant Breeding, 7(3): 490-496
In order to produce a good F1 hybrid variety in wheat, it is necessary to explore a new male-sterile cytoplasm and its nuclear restore gene(s). Four alloplasmic male sterile lines of wheat with Aegilops cytoplasm were developed to identify mitochondrial DNA (mtDNA) variation that could potentially be associated with cytoplasmic male sterility (CMS). mtDNA isolated from the Aegilops species, the respective male sterile lines and F1 hybrids were analyzed by RAPD markers. Reproducible polymorphisms were detected between the Aegilops species and male sterile lines. Above results indicated that mtDNA variation existed in the cytoplasm donors and male sterile lines resulted from genetic interactions between common wheat nucleus and Aegilops cytoplasm, and have affected the structure of the mitochondrial genome. Similar results were also obtained in male sterile lines and fertility-restored F1 hybrids. These demonstrated the variation of mtDNA in fertility restoration by the combination of the fertility restorer gene(s), and fertility restoration involved a strong influence of nuclear restorer genes on mtDNA organization. The variation of mtDNA in Aegilops species, their respective CMS lines and fertility-restored F1 hybrids may reflect the fertility divergence.
Cytoplasmic male sterility (CMS) is a very important trait for improving yield and quality of crops to meet the needs of increasing population in the world. In some well-studied species, the trait appears to be associated with mitochondrial DNA (mtDNA) mutations that disrupts mitochondrial function at a critical stage of anther development, thus causing male sterility (Hanson, 1991; Breiman and Galun, 1990; Mackenzie et al., 1994). Molecular studies between sterile and fertile plants have revealed variations in the restriction pattern of mtDNA and mitochondrial transcripts and proved that differences existed at the molecular level (Levings and Pring, 1976; Song and Hedgcoth, 1994). Alterations of specific mitochondrial genes and inserts have been identified (Song and Hedgcoth, 1994). Moreover, the combination of nucleus and cytoplasm from different plant species often leads to the CMS trait indicating that the sterility might probably be caused by functional incompatibility between the nuclear and mitochondrial genomes (Kofer et al., 1991). The nuclear loci suppressing the mitochondrial defects and restoring male fertility was reported as well (Hanson and Bentolila, 2004). The comparison of fertile and CMS plants provides a rare opportunity to examine the developmental regulation of mitochondrial function as well as the influence of nuclear background on mitochondrial gene expression. To identify the mtDNA sequences affecting the CMS trait, the general strategy was applied to compare cytoplasmic genomes in fertile and male sterile plants. Unfortunately, with a few exceptions, the comparative genomics strategy has mostly been unsuccessful (Hanson and Bentolila, 2004). mtDNA polymorphisms in CMS line and maintainer line often reflects the evolutionary divergence and have no correlation with CMS trait. Therefore, to compare recently diverged CMS and fertile plants become much more reliable for our researchers to identify CMS-associated gene regions. With this in view, the identification of genetic differences in mtDNA between fertile, male-sterile and restored fertile lines under nearly constant cytoplasmic background will reveal new strategies to locate CMS-associated gene regions.
Unlike the spontaneously occurred CMS that has been observed in some species, CMS in wheat often results from interspecific crosses, particularly the interspecific cytoplasm substitution. For example, the very famous T-type CMS wheat derived from the interspecific cross between Triticum timopheevi and Triticum aestivum. Such interspecific cytoplasmic substitutions often lead to incompatibility between the cytoplasm and the recipient nucleus, or results in cytoplasmic dysfunction (Leaver et al., 1988). Fertility restoration relies on the nuclear restoration gene(s) existing in Triticum timopheevi that suppress cytoplasmic dysfunction (Maan et al., 1984). However, the utility of T-type CMS system in hybrid production is limited for lack of new restorers and due to wrinkled grain shape and pre-harvest sprouting of developed hybrids. Besides, the application of wheat heterosis is also impeded by other factors, including low hybrid seed production rate, difficulty in maintaining purity of male sterile lines, high seeding rate etc.. In order to develop a good F1 hybrid variety in wheat, it is necessary to explore a new male-sterile cytoplasm and its nuclear restore gene(s). With this in view, we developed a series of alloplasmic male sterile lines (or called nuclear-cytoplasm substitution lines) of wheat with Aegilops cytoplasm by recurrently backcrossing Ae. kotschyi-Chris with a recurrent parent 90-110 (Triticum aestivum). In comparison with T-type CMS system, our male sterile lines have some significant advantages, such as the abundance of restorers, the characteristic of easy-restoration-and-maintanence, and round grain shape of hybrids, so it is being regarded as the most promising among all alien cytoplasms for hybrid wheat production (Zhang and Yang, 1989; Zhang, 1992).
In this paper, an attempt was made to study mtDNA variation using RAPD (random amplified polymorphic DNA) markers on the Aegilops species, male sterile lines and fertility-restored F1 hybrids under a nearly constant cytoplasmic background.
1 Materials and methods
1.1 Plant materials
Four Aegilops species viz. Ae. kotschyi, Ae. variabilis, Ae. ventricosa and Ae. bicornis, and two fertile common wheat varieties namely 90-110 and 5253, were used in the present investigation. Four male sterile lines viz. ms (Ae. kotschyi)-90-110, ms (Ae. variabilis)-90-110, ms (Ae. ventricosa)-90-110 and ms (Ae. bicornis)-90-110 were derived from recurrent backcross of Ae. kotschyi-Chris, Ae. variabilis-Chris, Ae. ventricosa-Chris and Ae. bicornis-Chris to 90-110 respectively, for more than 20 generations. The wheat variety 90-110 was used as the maintainer line while the variety 5253 was used as the restorer line to restore fertility of the four male sterile lines.
This study was carried out with emphasis on mitochondrial DNA variation among the Aegilops species, their respective CMS lines and fertility-restored F1 hybrids. Because the cytoplasmic genomes are maternally inherited in wheat and its relatives, the plant materials used for isolation of mitochondrial DNA are under nearly constant cytoplasmic background (Table 1).
|
|
1.2 DNA isolation
mtDNA was isolated from etiolated shoots as described by Li et al. (2007). The protocol includes: mitochondria isolation with differential centrifugation, DNase treatment, lysis with SDS and proteinase K, removing protein by TE-saturated phenol/chloroform extraction and a final RNase A treatment for obtaining mtDNA. The mtDNA samples were tested for purity using spectrophotometry, agarose gel electrophoresis and restriction enzyme digestions. It was proved that the mtDNA was not contaminated by nuclear DNA, plastid DNA, RNA and protein, and was successfully used for PCR, cloning and southern blot analyses. Total DNA was isolated following CTAB method as described by Murray and Thompson (1980).
1.3 DNA amplification by RAPD
RAPD amplification of mtDNA and total DNA was performed following Williams et al. (1990). Ten-nucleotide primers of arbitrary sequence were obtained from Operon Technologies and Shanghai Sangon Biological Engineering Technology. Amplification reactions were performed in total volumes of 25 μL containing 10 mmol/L Tris-HCl (pH 8.0), 50 mmol/L KCl, 2 mmol/L MgCl2, 100 μmol/L of dNTPs, 0.4 μmol/L primer, 25 ng mtDNA or total DNA as template, and 1.5 units Taq DNA polymerase. Amplification was performed in a Bio-Rad MyCycler Thermal Cycler programmed with an initial denaturation step for 3 min at 95℃ followed by 40 cycles of denaturation for 1 min at 94℃, annealing for 1 min at 37℃, primer extension for 2 min at 72℃, and a final extension step of 5 min at 72℃. Amplification products were analyzed by gel electrophoresis in 1.5% agarose gel and detected by staining with ethidium bromide. Amplification patterns were analyzed by Gene Tools from Syngene to evaluate molecular weight, and all the RAPD amplification reactions were repeated more than three times.
2 Results
2.1 RAPD analysis of Aegilops species and their respective CMS lines
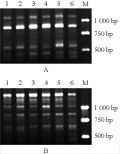
A total of 120 primers were used for screening RAPD markers to distinguish the Aegilops species, their respective CMS lines and fertility-restored F1 hybrids using mtDNA extracted from etiolated shoots (see supporting online material). The primer sets showing stable polymorphisms are listed in table 2. Of therm, eight primers were found showing stable polymorphism in the Aegilops species and their respective male sterile lines. In Ae. kotschyi, primer OPY-01 produced a 600 bp band while in the male sterile line ms (Ae. kotschyi)-90-110 a 580 bp band was observed (Figure 1A).In Ae. variabilis, primer S32 can amplify a 1 770 bp band while in ms (Ae. kotschyi)-90-110 the primer can amplify 1 700 bp and 1 440 bp bands (Figure 1B). In Ae. ventricosa, primer S22 can amplify a 2 310 bp band, while in ms (Ae. ventricosa)-90-110 the primer can amplify a 1 770 bp band (Figure 1C).
|
|
For Ae. bicornis and ms (Ae. bicornis)-90-110 different fingerprint patterns were obtained based on amplification by primer pairs S202, OPB-05, OPA-04, S153 and S21, respectively. In ms (Ae. bicornis)-90-110, primer S202 can amplify a specific 2 500 bp band while in Ae. bicornis the 2 500 bp band can not be amplified (Figure 2A). Also in ms (Ae. bicornis)-90-110, primer OPB-05 can amplify a specific 3 020 bp band, while in Ae. bicornis the 3 020 bp band can not be amplified (Figure 2B). In Ae. bicornis, primer OPA-04 can amplify a specific 1 000 bp band while in ms (Ae. bicornis)-90-110 the 1 000 bp band can not be amplified (Figure 2C). Primer S153 can amplify a 2 270 bp band in ms (Ae. bicornis)-90-110, but in Ae. bicornis the specific band was absent (Figure 2D). Similarly, premer S21 can amplify a 2 300 bp band in ms (Ae. bicornis)-90-110 while in Ae. bicornis the specific band was absent (Figure 2E). In the CMS lines (nuclear-cytoplasm substitution lines), nuclear genome of the Aegilops was substituted in the successive backcross with common wheat 90-110. Above results indicate that mtDNA variation in the donors of cytoplasms and CMS lines result from genetic interactions between 90-110 nuclear genes and Aegilops cytoplasm, and have affected the structure of the mitochondrial genome. The structural alternations of the mitochondrial genome are possible factors to cause the mitochondrial defect and CMS.
|
|
|
|
2.2 RAPD analysis of CMS lines and fertility-restored hybrids
To assess the effect of fertility restoration on mitochondrial genome, CMS plants were crossed with the restorer line of common wheat 5253 to obtain fertile F1 hybrids. Subsequently, remarkable differences were found in the amplification patterns of mtDNA between CMS lines and fertile F1 hybrids. Of the 120 RAPD primers used, four primers OPY-01, OPD-05, S21 and OPP-02 showed stable polymorphisms in the male sterile lines and their respective fertility-restored F1 hybrids. In male sterile line ms (Ae. kotschyi)-90-110, primer OPY-01 can amplify a specific 580 bp band while in ms (Ae. kotschyi)-90-110×5253 (fertility-restored hybrids) the primer can amplify a specific 550 bp band (Figure 1A). In ms (Ae. variabilis)-90-110, primer OPD-05 can amplify a specific 2 200 bp band, while in ms (Ae. variabilis)-90-110×5253 the band was absent (Figure 3A). In ms (Ae. ventricosa)-90-110×5253, primer S21 can amplify two specific bands of 2 020 bp and 1 610 bp, respectively, while in male sterile line ms (Ae. ventricosa)-90-110 the spceific bands were absent (Figure 3B). Similarly, in ms (Ae. bicornis)-90-110×5253, primer OPP-02 can amplify a 2 530 bp band while in ms (Ae. bicornis)-90-110 the specific band was absent (Figure 3C). These results suggested that the mitochondrial genomes were altered in fertility restoration by the combination of fertility restorer gene(s), and fertility restoration involving a strong influence of nuclear restorer genes on mtDNA organization. Variation of mitochondrial genome might be dependent on different nuclear backgrounds.
|
|
To verify the observed polymorphisms are indeed derived from the mitochondrial DNA, nuclear DNA from the CMS lines and fertility-restored F1 hybrids was isolated and amplified using the polymorphic primers OPY-01 and OPD-05, respectively. Results showed that the amplified bands were same between CMS lines and F1 hybrids. Primer OPY-01 showed no polymorphism in male sterile line ms (Ae. kotschyi)-90-110 and its fertility-restored hybrid ms (Ae. kotschyi)-90-110×5253 (Figure 4A), and the bands amplified by primer OPD-05 was also indistinguishable between ms (Ae. variabilis)-90-110 and ms (Ae. variabilis)-90-110×5253 (Figure 4B).
|
|
3 Discussion
It is now widely believed that one of the cause of cytoplasmic male sterility (CMS) is the functional incompatibility between the nuclear and mitochondrial genome. Mitochondrial mutations, such as multiple intragenic recombination and insertion of sequences of unknown origin, are associated with CMS (Dewey et al., 1986; Young and Hanson, 1987; Mackenzie et al., 1988). In most cases, CMS is associated with a variety of mitochondrial DNA rearrangements, and no two CMS mutations described to date have been identical (Sandhu et al., 2007).
In the present experiment the combination of Triticum aestivum nuclei and alien Aegilops cytoplasm resulted in the genetic interactions or functional incompatibility between the nuclear and mitochondrial genomes. This may cause some mutations in the mitochondrial genome or the expression of some genes may get suppressed due to the nucleus-mitochondrion incompatibility. Thus, mitochondrial mutations induce abnormal anther development and the CMS trait occurred. The introduction of nuclear restorer gene(s) from restorer line 5253 reversed the changes caused to the mitochondrial genome thereby resulting in fertile F1 hybrids. The results confirmed the mtDNA variation among Aegilops species, CMS lines and their fertility-restored F1 hybrids.
In theory, the cytoplasms of these four CMS lines are derived from the respective Aegilops species (Ae. kotschyi, Ae. variabilis, Ae. ventricosa and Ae. bicornis). However, distinct differences were detected between Aegilops and CMS lines. The variation of mtDNA in Aegilops spp. and their respective CMS lines may reflect the fertility divergence. Since a variety of mtDNA mutations can cause the CMS trait, it was not possible to determine which variation of mtDNA patterns is linked with the CMS trait. Further studies, are therefore needed to reveal the relationship between the polymorphic markers and CMS trait.
In most cases, fertility restorer genes appear to regulate gene expression at a transcriptional or posttranscriptional level (Singh et al., 1996; Li et al., 1998). Recent studies also have shown evidence of fertility restoration through mtDNA alterations. In maize, spontaneous reversion to fertility in CMS-S maize involves alterations of the mitochonrial genome (Schardl et al., 1985). In common bean, fertility restorer gene Fr locus results in the loss of a particular CMS-associated mtDNA region (He et al., 1995). Our results demonstrated the variation of mtDNA in CMS lines and their fertility-restored F1 hybrids. It's possible that fertility restoration involves a strong influence of the restorer genes on mtDNA organization. Although the differences in RAPD patterns are not necessarily associated with CMS trait, they can be informative molecular markers to map CMS-associated gene regions. Further studies are under way converting the RAPD markers into SCAR markers and cloning CMS-associated mitochondrial gene.
Acknowledgements
We appreciate the efforts taken by Mr T.V. Murali in revising the english of the manuscript. This research was supported by National Nature Science Foundation of China (301705760), Chinese National Programs for High Technology Research and Development (2002AA207004) and Special Program of National Agricultural Biotechnology & Breeding Center Yangling Branch.
References
Breiman A., and Galun E., 1990, Nuclear-mitochondrial interrelation in angiosperms, Plant Science, 71: 3-19
Dewey R.E., Levings C.S., and Timothy D.H., 1986, Novel recombinations in the maize mitochondrial genome produce a unique transcriptional unit in the Texas male-sterile cytoplasm, Cell, 44(3): 439-449 doi:10.1016/0168-9452(90)90063-T
Hanson M.R., 1991, Plant mitochondrial mutations and male sterility, Annu. Rev. Genet., 25: 461-486
Hanson M.R., and Bentolila S., 2004, Interactions of mitochondrial and nuclear genes that affect male gametophyte development, The Plant Cell, 16: S154-S169 doi:10.1146/annurev.ge.25.120191.002333
He S., Yu Z.H., Vallejos C.E., and Mackenzie S.A., 1995, Pollen fertility restoration by nuclear gene Fr in CMS common bean: an Fr linkage map and the mode of Fr action, Theor. Appl. Genet., 90(7-8): 1056-1062 doi:10.1105/tpc.015966
Kofer W., Glimelius K., and Bonnett H.T., 1991, Modifications of mitochondrial DNA cause changes in floral development in homeotic-like mutants of tobacco, The Plant Cell, 3(8): 759-769 doi:10.1007/BF00222921
Leaver C.J., Isaac P.G., Small I.D., Bailey-Serres J., Liddell A.D., and Hawkesford M.J., 1988, Mitochondrial genome diversity and cytoplasmic male sterility in higher plants, Philos. Trans. R. Soc. Lond. B., 319(1193): 165-176 doi:10.1098/rstb.1988.0040
Levings C.S., and Pring D.R., 1976, Restriction endonuclease analysis of mitochondrial DNA from normal and Texas cytoplasmic male-sterile maize, Science, 193(4248): 158-160 doi:10.1126/science.193.4248.158
Li W.Q., Zhang G.S., Wang K., Niu N., and Pan D.L., 2007, An efficient method for isolation of mitochondrial DNA in wheat, Yichuan (Hereditas), 29(6): 771-775
Li X.Q., Jean M., Landry B.S., and Brown G.G., 1998, Restorer genes for different forms of Brassica cytoplasmic male sterility map to a single nuclear locus that modifies transcripts of several mitochondrial genes, Proc. Natl. Acad. Sci., USA, 95(17): 10032-10037 doi:10.1073/pnas.95.17.10032
Maan S.S., Lucken K.A., and Bravo J.M., 1984, Genetic analyses of male-feritility restoration in wheat. â… : Chromosomal location of Rf genes, Crop Science, 24: 17-20 doi:10.2135/cropsci1984.0011183X002400010005x
Mackenzie S., He S., and Lyznik A., 1994, The elusive plant mitochondrion as a genetic system, Plant Physiology, 105(3): 775-780
Mackenzie S.A., Pring D.R., Bassett M.J., and Chase C.D., 1988, Mitochondrial DNA rearrangement associated with fertility restoration and cytoplasmic reversion to fertility in cytoplasmic male sterile Phaseolus vulgaris L., Proc. Natl. Acad. Sci., USA, 85(8): 2714-2717 doi:10.1073/pnas.85.8.2714
Murray M.G., and Thompson W.F., 1980, Rapid isolation of high molecular weight plant DNA, Nucleic Acids Research, 8(19): 4321-4325 doi:10.1093/nar/8.19.4321
Sandhu A.P.S., Abdelnoor R.V., and Mackenzie S.A., 2007, Transgenic induction of mitochondrial rearrangements for cytoplasmic male sterility in crop plants, Proc. Natl. Acad. Sci., USA, 104(6): 1766-1770 doi:10.1073/pnas.0609344104
Schardl C.L., Pring D.R., and Lonsdale D.M., 1985, Mitochondrial DNA rearrangements associated with fertile revertants of S-type male-sterile maize, Cell, 43(1): 361-368
Singh M., Hamel N., Menassa R., Li X.Q., Young B., Jean M., Landry B.S., and Brown G.G., 1996, Nuclear genes associated with a single Brassica CMS restorer locus influence transcripts of three different mitochondrial gene regions, Genetics, 143(1): 505-516
Song J., and Hedgcoth C., 1994, Influence of nuclear background on transcription of a chimeric gene (orf256) and coxI in fertile and cytoplasmic male sterile wheats, Genome, 37(2): 203-209 doi:10.1139/g94-028
Williams J.G., Kubelik A.R., Livak K.J., Rafalski J.A., and Tingey S.V., 1990, DNA p olymorphisms amplified by arbitrary primers are useful as genetic markers, Nucleic Acids Research, 18(22): 6531-6535 doi:10.1093/nar/18.22.6531
Young E.G., and Hanson M.R., 1987, A fused mitochondrial gene associated with cytoplasmic male sterility is developmentally regulated, Cell, 50(1): 41-49
Zhang G.S., 1992, Studies of genetic mechanism of haploid induced by male sterile lines of 1B/1R wheat with Ae. kotschyi, Ae. variabilis cytoplasms and its performance of fertility restoration, Yichuan Xuebao (Acta Genetica Sinica), 19(3): 266-277
Zhang G.S., and Yang T.Z., 1989, A preliminary on the male sterile lines of wheat with Ae. ventricosa, Ae. kotschyi and Ae. vauiabilis cytoplasms, Zuowu Xuebao (Acta Agronomica Sinica), 15(1): 1-10
. PDF(382KB)
. FPDF(win)
. HTML
. Online fPDF
Associated material
. Readers' comments
Other articles by authors
. Wenqiang Li
. gaisheng zhang
. Na Niu
. Fang Wei
. Kui Wang
. Dongliang Pan
Related articles
. Aegilops
. Wheat
. Cytoplasmic male sterility
. mtDNA
. RAPD markers
Tools
. Email to a friend
. Post a comment
.png)